Mar 28 2013
Remembering the Memorable
One of the features of any network is the appearance of motifs, patterns that recur within a network much more often than expected via any sort of random occurence. The first four notes of Beethoven’s Fifth Symphony are an example of a motif: This opening phrase is one of the most widely recognized in music. It has mystified musicians, historians and philosophers for 200 years. Music critic Matthew Guerrieri says it’s “short enough to remember and portentous enough to be memorable.”
These small circuits can be considered as simple building blocks from which the network is composed. This analogy is quite useful, since many of these motifs would appear to have their corollaries in electronic circuitry. Motifs appear to play an important role in transcription factor regulation, providing the simple computational elements that function as integrated wholes to produce complex algorithmic outcomes.
Like the digital circuitry in your computer, clusters of network motifs are capable of computational processes. Think about this: Humans share about 98% of their genome (at least the sequences) with apes. This of course begs the question ‘why are we not more similar?’ The answer is that while we share much of the base sequences, there are tremendous differences in the computational knowledge that acts upon these sequences, in particular the networks involved in transcription factor regulation: operons, regulons and modulons. It is the combinatoric wisdom that seems to differentiate between the classes of life forms. This is especially true with regard to gene regulatory elements, which lie within the 98% of the DNA that does not contain gene coding. Gene regulatory elements instruct genes as to when, where and at what levels to turn on or off. [1]
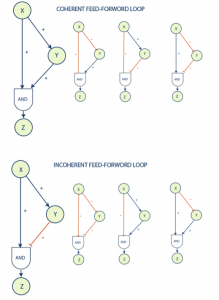
One of the most common and interesting motifs found in biological systems is known as a feed-forward loop. This motif is commonly found in many gene systems and organisms. The motif consists of three genes and three regulatory interactions. The target gene Z is regulated by 2 transcription factors X and Y and in addition TF Y is also regulated by transcription factor X. The target gene is usually operated on in a logical AND manner, in that it requires both inputs to be logically true (both X and Y are required for Z activation) in order to activate. Since each of the regulatory interactions may either be stimulatory or inhibitive there are possibly eight types of FFL motifs.
Feed-forward loops are classified as coherent or incoherent. A coherent feed-forward loop is distinguished by the fact that the final actions of transcription factors X and Y are symmetrical; i.e. they result in the same type of stimulus (stimulation-stimulation or inhibition-inhibition) at their termination, Z. Incoherent feed-forward loops result in differing signals (stimulus-inhibition) at their termination.
The coherent type 1 feed-forward loop (C1-FFL) with an AND gate was shown to have a function of a ‘sign-sensitive delay’ element and a persistence detector both theoretically and experimentally with the arabinose system of E. coli. [2] This means that this motif can provide pulse filtration in which short pulses of signal will not generate a response but persistent signals will generate a response after short delay. The shut off of the output when a persistent pulse is ended will be fast.
The incoherent type 1 feed-forward loop (I1-FFL) is a pulse generator and response accelerator. The two signal pathways of the I1-FFL act in opposite directions where one pathway activates Z and the other represses it. When the repression is complete this leads to a pulse-like dynamics. I1-FFL is a pulse generator and response accelerator. [3] In some cases the same regulators X and Y regulate several Z genes of the same system. This is known as a multi-output feed-forward loop. By adjusting the strength of the interactions this motif was shown to determine the temporal order of gene activation. [4]
Other common motifs include auto-regulation, single input modules and dense overlapping regulons (DOR). Negative auto-regulation (NAR) occurs when a transcription factor represses its own transcription. This motif was shown to perform two important functions. The first function is response acceleration. NAR was shown to speed-up the response to signals both theoretically and experimentally. The second function is increased stability of the auto-regulated gene product concentration against stochastic noise, thus reducing variations in protein levels between different cells. [5] Positive auto-regulation (PAR) occurs when a transcription factor enhances its own rate of production. Opposite to the NAR motif this motif slows the response time compared to simple regulation. In the case of a strong PAR the motif may lead to a bimodal distribution of protein levels in cell populations. [6] The single input module (SIM) motif occurs when a single regulator regulates a set of genes with no additional regulation. This is useful when the genes are cooperatively carrying out a specific function and therefore always need to be activated in a synchronized manner. In the dense overlapping regulon motif, many inputs regulate many outputs. This motif occurs in the case that several regulators combinatorially control a set of genes with diverse regulatory combinations. This motif was found in E. coli in various systems such as carbon utilization, anaerobic growth, stress response and others. [4]
Coding for motif detection in Datapunk QUODLIBET was quit challenging. Although there are many helpful modules in the CPAN archives, there exists no simple module for FFL motif detection in network graphs. However, with experimentation I was able to work out a simple algorithm. Any metabolic map can display FFLs (if there are any in them) by clicking the FFL icon in the floating tool bar (indicated by red arrow).
Here we see a Type 2 Coherent FFL motif in the Datapunk Adipocytokine map. Suppressor of cytokine signaling (SOCS) proteins are key regulators of immune responses and exert their effects in a classic negative-feedback loop. SOCS3 is transiently expressed by multiple cell lineages within the immune system and functions predominantly as a negative regulator of the leptin receptor and downstream cytokines that activate Janus Kinase 2.
To me, looking for basic circuitry motifs in metabolic maps (and understanding their function and significance) is equivalent to a composer who hears an entire symphonic arrangement in his head while reading a musical score, while the rest of us mere mortals just see black dots on white paper.
FOOTNOTES
1. Davidson E. The Regulatory Genome. Academic Press. (2006)
2. Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks.J Mol Biol. 2003 Nov 21;334(2):197-204
3. Mangan S, Itzkovitz S, Zaslaver A, Alon U.The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol. 2006 Mar 10;356(5):1073-81. Epub 2005 Dec 19.
4. Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman and Hall/ CRC. New York, NY (2007)
5. Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323 (5): 785–93. (November 2002)
6. Maeda YT, Sano M. Regulatory dynamics of synthetic gene networks with positive feedback. J. Mol. Biol. 359 (4): 1107–24. (June 2006)